Abstract
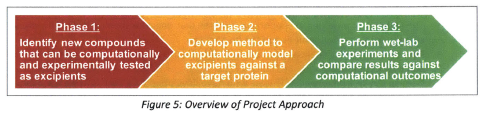
Computational modelling has completely redefined the experimentation process in many industries, allowing large sets of design concepts to be tested quickly and cheaply very early in the innovation process. Harnessing the power of computational modelling for protein drug formulation has numerous, currently unrealized, benefits. This project aims to be the first step in the development of a high throughput predictive computational model to screen for excipients that would decrease protein aggregation in solution and thus increase its stability and enable clinical effectiveness. Protein drug formulation currently relies heavily on empirical evidence from wet-lab experiments and personal experience. During the biologic drug development process, proteins that target specific disease pathways are identified, developed, isolated, and purified. Scientists then conduct a series of wet-lab experiments to identify the optimal formulation that will allow the protein to be used as a drug therapy. A critical part of formulation development is the identification of inactive ingredients called excipients that perform various important functions including prevention of protein aggregation. Despite their critical role in enabling proteins to be effective therapies, very little is understood about excipient-protein interaction. Furthermore, often a limited set of compounds are tested for their use as excipients since wet-lab experiments are expensive and time consuming. This project accomplishes the following goals: ** Identification of databases of compounds that could be used as excipients in biologic formulation; ** Development of a high throughput method to computationally model a target protein and 247 potential excipients; ** Evaluation of potential relationship between computational output and wet-lab results based onxperimentation with 32 of the 247 excipients; ** Recommendations on next steps that include feedback on types of proteins and excipients to be tested for the validation of the method developed in this project.
URI: http://hdl.handle.net/1721.1/112488